AMAES: Augmented Masked Autoencoder Pretraining on Public Brain MRI Data for 3D-Native Segmentation
* Equal Contribution
Data & model publicly available.

This study investigates the impact of self-supervised pretraining of 3D semantic segmentation models on a large-scale, domain-specific dataset. We introduce 🧠BRAINS-45K, a dataset of 44,756 brain MRI volumes from public sources, the largest public dataset available, and revisit a number of design choices for pretraining modern segmentation architectures by simplifying and optimizing state-of-the-art methods, and combining them with a novel augmentation strategy. The resulting AMAES framework is based on masked-image-modeling and intensity-based augmentation reversal and balances memory usage, runtime, and finetuning performance. Using the popular U-Net and the recent MedNeXt architecture as backbones, we evaluate the effect of pretraining on three challenging downstream tasks, covering single-sequence, low-resource settings, and out-of-domain generalization. The results highlight that pretraining on the proposed dataset with AMAES significantly improves segmentation performance in the majority of evaluated cases, and that it is beneficial to pretrain the model with augmentations, despite pretraing on a large-scale dataset.
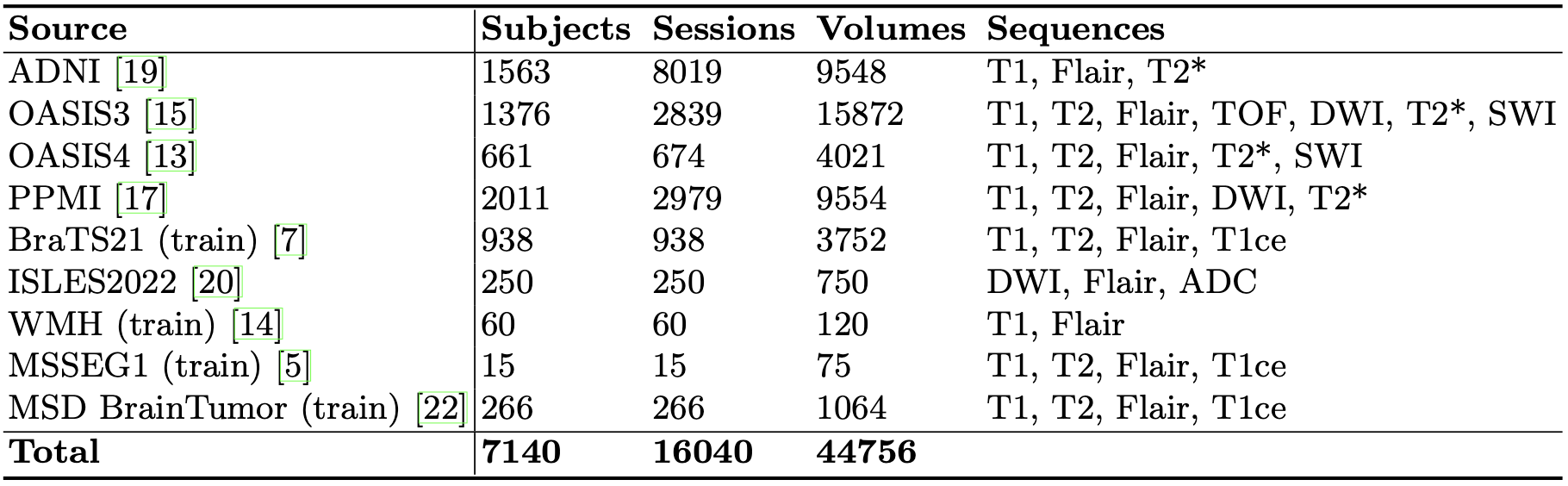
A dataset of 44.756 Brain MRI volumes collected from public sources, featuring a diverse set of acquisition parameters and patient populations. It is a compilation of data from four large non-labelled datasets (ADNI, OASIS3, OASIS4, PPMI) and five challenge datasets (MSD, BraTS21, ISLES22, WMH, MSSEG1). The resulting dataset is highly heterogeneous and assembled to simulate the characteristics of clinical data. Overview of the dataset can be found in the table below. The dataset includes data acquired at both 1.5T and 3T.
Preprocessing. To compile the diverse data into a suitable pretraining dataset, we transformed all volumes to the RAS coordinates system, resampled to isotropic 1mm spacing, clipped values to the 99% percentile, and z-normalized on a volume level. Afterwards, each volume is cropped to the minimum bounding box of the brain. The preprocessing pipeline is based on the Yucca framework for medical imaging. Code to reproduce the final version of the dataset can be found here. Usage of the dataset needs to comply with the individual legal requirements of each source.
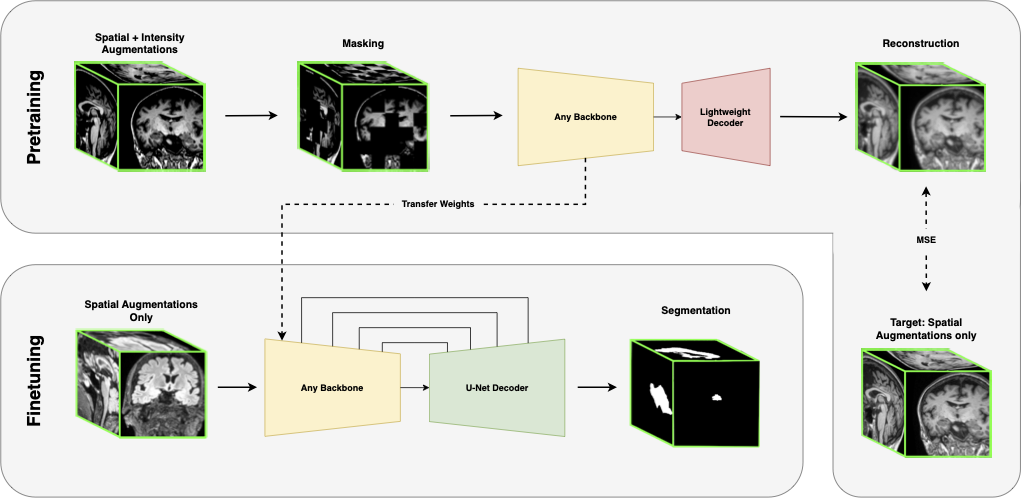
During pretraining, spatial and intensity-based augmentations are applied to an image patch. The patch is masked and passed through the model, which consists of a backbone encoder and a lightweight decoder, to reconstruct the image. The reconstruction target is the unmasked image, with only spatial transformations applied. During finetuning, only spatial augmentations are applied to the input. The backbone encoder weights are transferred, while a new U-Net decoder is initialized. Skip connections are only used during finetuning.
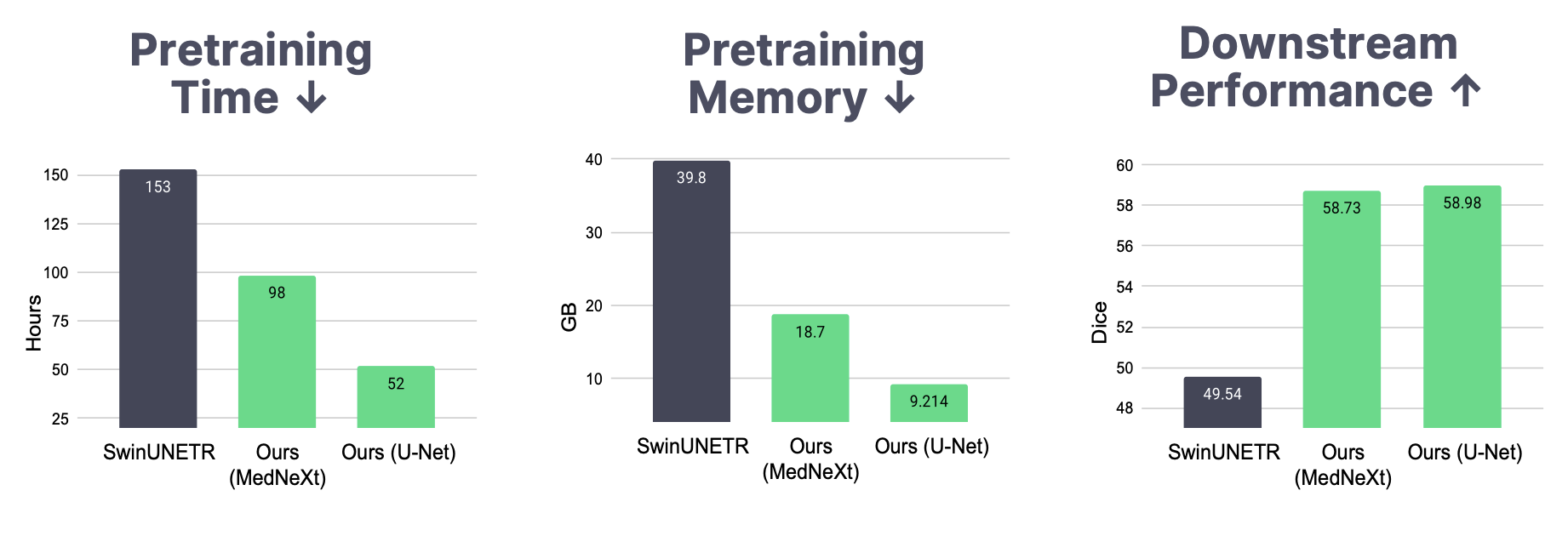
We evaluate AMAES in a setup designed to test the capacity of pretrained models by restricting the amount of training data during finetuning to 20 labeled training examples. The evaluation is then performed along two dimensions: (i) How does AMAES compare to the pretrained SwinUNETR?, and (ii) What is the impact of pretraining on downstream performance?
To evaluate (i), we pretrain SwinUNETR with the exact configuration given in on 🧠BRAINS-45K, which includes both a contrastive loss, a rotation loss and a reconstruction loss, as well as a different choice of masking ratio and mask size. To ensure a fair comparison, SwinUNETR is pretrained for 100 epochs, similar to AMAES, and is pretrained with patch size 1283, instead of patch size 963 and 90 epochs.
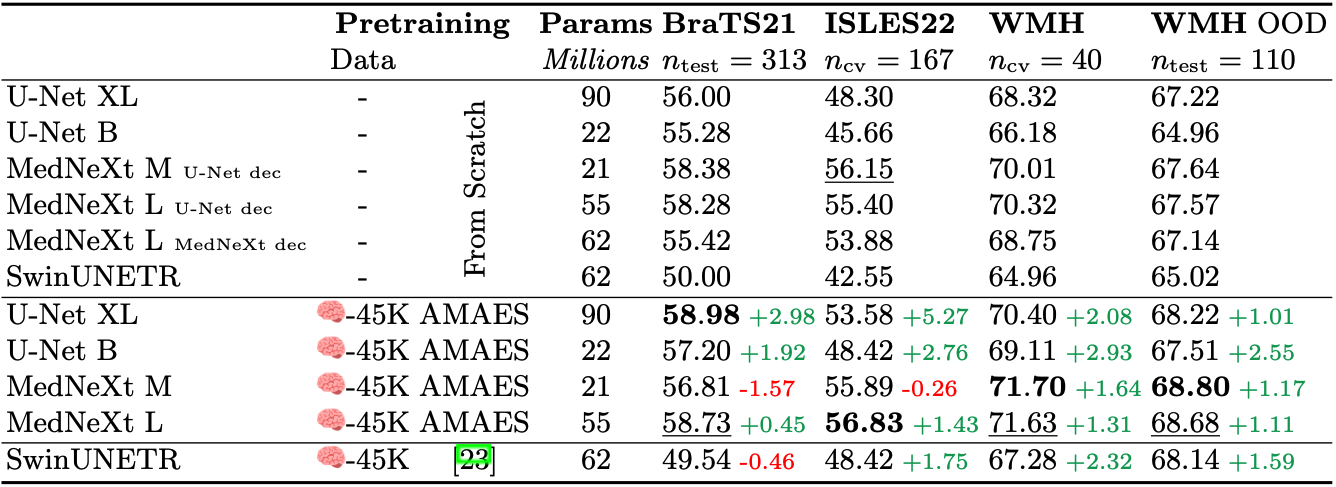
To address (ii), we apply AMAES to a set of convolutional backbones and evaluate the difference between training from scratch and finetuning the pretrained model. The backbone models include a U-Net in two sizes: XL (90M parameters) and B. The U-Net B is very similar in size to the one used in nnUNet (22M parameters, when trained with the default setting max vram of 12 GB). Further, we explore using the modernized U-Net architecture MedNeXt, which uses depth-wise seperable convolution to introduce compound scaleable medical segmentation models. All MedNeXt models are trained with kernel size 3 and do not use UpKern. The networks are finetuned and trained from scratch using the same hyperparameters. Models trained from scratch use the full augmentation pipeline. Results for both (i) and (ii) are given in Table 2.
@article{munk2024amaes,
title={AMAES: Augmented Masked Autoencoder Pretraining on Public Brain MRI Data for 3D-Native Segmentation},
author={Munk, Asbjørn and Ambsdorf, Jakob and Llambias, Sebastian and Nielsen, Mads},
journal={arXiv preprint arXiv:2408.00640},
year={2024}
}